In vitro Screening and Identification of P-Solubilizing Rhizobacteria Associated with Sorghum bicolor L.
Journal of Agriculture Research- Juniper Publishers
Abstract
In the present study, P-solubilizing rhizobacteria were screened and identified from Sorghum bicolor
L root adhering soil and root which were collected from sorghum growing
zones of Tigray, Ethiopia. A total of 94 bacteria were isolated from
root washing solutions and surface sterilized roots. These isolates were
evaluated for their ability to solubilize phosphates on Pikovskaya’s
agar plates. The P-solubilizing bacterial isolates were identified by
GEN III Biolog bacterial identification system. Fifty four of the 94
(57.5%) rhizobacterial isolates showed clearly visible haloes
(>0.50cm) around their colonies on Pikovskaya’s agar after seven days
of incubation. The solubilization index (SI) of the potential
P-solubilizing rhizobacterial isolates differed significantly
(p<0.05) and ranged from 0.5 to 4.83. Gram negative rhizobacteria
dominated the identified P-solubilizing Rhizobacteria isolates and
produced larger solubilization indices when compared with the
Gram-positive isolates. Members of the phosphobacteria were dominated by
the genus Pseudomonas (35.71%). Some of the isolates lost their
capacity for phosphate solubilization on repeated sub-culturing.
Overall, this finding indicated that there is a great number of
rhizobacterial potential associated with Sorghum bicolor L which can be utilized for development of P-solubilizing bio-fertilizers.
Keywords: P-solubilizing rhizobacteria; Sorghum bicolor L.; Biolog bacterial identification
Abbreviations:
PGPR: Plant Growth Promoting Rhizobacteria; PSM: Phosphate Solubilizing
Microbes; PSB: Phosphate Solubilizing Bacteria; CD: Colony Diameter;
SI: Solubilization Index; BUG: Biolog Universal Growth
Introduction
Plant growth promoting rhizobacteria (PGPR) flourish
in the rhizosphere of plant, which may grow in, on, or around plant
tissues and exert beneficial effects on plant development [1,2]. They
possess the capacity to stimulate plant growth either directly or
indirectly [3]. PGPR can affect plant growth by a wide range of
mechanisms such as solubilization of inorganic phosphate, production of
phyto-hormones, siderophores and organic acids, lowering of plant
ethylene levels, N2 fixation and bio-control of plant diseases [4,5].
The use of such beneficial bacteria as bio-fertilisers and bio-control
agents has currently attracted increased interest world-wide in attempts
to achieve sustainability, particularly in agriculture, forestry and
horticulture [5].
The number of PGPR that have been identified has seen
a great increase in the last few years, mainly because of the role of
the rhizosphere as an ecosystem has gained importance in the functioning
of the biosphere. Various species of bacteria like Pseudomonas, Azospirillum, Azotobacter, Klebsiella, Enterobacter, Alcaligenes, Arthrobacter, Burkholderia, Bacillus and Serratia
have been reported to enhance plant growth. There are several
PGPR inoculants currently commercialized that seem to promote growth
through at least one mechanism; suppression of plant disease (termed
Bio-protectants), improved nutrient acquisition (Bio-fertilizers), or
phyto-hormone production (Bio-stimulants) [2].
The use of PGPR offers an attractive way to replace
chemical fertilizer, pesticides, and supplements; most of the isolates
result in a significant increase in plant height, root length, and dry
matter production of shoot and root of plants. The economic and
ecological problems of today have re-invigorated the idea of using
bio-fertilizers and bio-control agents in order to reduce the
application of costly and environmentally-polluting agrochemicals to a
minimum [6,7]. Agrochemicals (namely fertilizers and pesticides) have
greatly influenced natural rhizosphere microbes in agro-systems [8].
Plant beneficial microbial bio-resources promise to replace or
supplement many such destructive, high intensity practices and support
ecofriendly crop production [6,7]. In particular, plant growth promoting
rhizobacteria (PGPR) for the benefits of agriculture and ecosystem
functions is gaining worldwide importance and acceptance [6,7,9,10].
Phosphorus is the second most important nutrient for plants,
after nitrogen. It exists in soil as mineral salts or incorporated
into organic compounds. Despite these phosphorus compounds
being abundant in agricultural soils, the majority of them occur
in an insoluble form. Plants require approximately 30μmol l-1
of phosphorus for maximum productivity, but only about 1μmol
l-1 is available in many soils. Therefore, the unavailability of
phosphorus in many soils has been recognized as a major growth
limiting factor in agricultural and horticultural systems. This
necessitates the application of soluble forms of phosphorus in
the form of phosphate fertilizers, which in itself has constraints in
that it too is rapidly immobilized (fixed) to insoluble forms upon
its application in the soil due to its reaction with aluminum and
iron minerals. The efficiency of applied phosphorus rarely exceeds
30% due to fixation in soil. It is also lost as a result of run-off and
leaching, leaving as little as 10-20% available for plant utilization.
Phosphate fertilizers are dependent on phosphorus derived from
phosphate rock, which is a non-renewable resource and current
global reserves may be depleted in 50-100 years. Therefore,
exploring alternative forms of agriculture, where nutrient
conservation is key, is of vital importance [11].
Several reports have indicated that different bacterial species,
particularly rhizosphere colonizing bacteria, have the ability to
liberate organic phosphates or to solubilize insoluble inorganic
phosphate compounds such as tri-calcium phosphate, di-calcium
phosphate, hydroxyapatite, and rock phosphate. These bacteria
make available the soluble phosphates to the plants, and in return
gain root borne carbon compounds, mainly sugars and organic
acids, necessary for bacterial growth [12]. Current research
suggests that the inoculation of crops with Phosphate Solubilizing
Microbes (PSM) has the potential to reduce application rates of
phosphate fertilizer by 50% without significantly reducing crop
yield [13,14]. Phosphate Solubilizing Bacteria (PSB) may also
be useful in the phyto-remediation of heavy metal impacted soil
[15,16] or for bioleaching of rare Earth elements for mined ores
[17].
Most soils in tropical and subtropical areas are predominantly
acidic and extremely P-deficient due to their strong fixation of
P as insoluble phosphates of iron and aluminum [9,12,18]. This
leads to wide P deficiency which is particularly the case for the
large parts of Ethiopian soils [19,20]. To alleviate P deficiency,
chemical phosphate fertilizers are widely used. However, a large
proportion of the soluble forms of P fertilizers is precipitated in
insoluble form soon after application and becomes unavailable
to plants [21]. This in turn leads to a need for excessive and
repeated application of soluble P fertilizers, which in addition to
the economic constraint can pose a serious threat to groundwater.
These have been the major stresses that constrain the production
of crops in the country.
Thus, in relation to this fact, P-solubilizing Rhizobacteria
associated with cultivated Sorghum plant roots that displayed
bio-fertilizer characteristics and have potential applications as
native P-solubilizing bacterial bio-fertilizers were screened and
identified in this study.
Materials and Methods
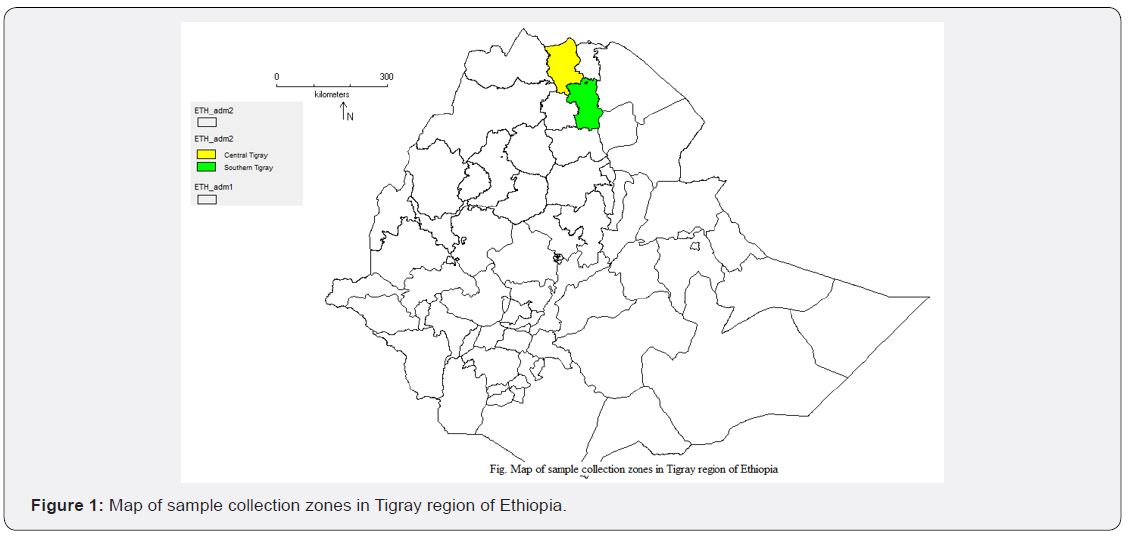
Description of sample collection areas
Sample collection was carried out in two major sorghum
producing zones of Tigray region in Ethiopia. The sample
collection site is shown in Figure 1. It comprises Central Tigray
and South Tigray zones which are found in the northern part of
Ethiopia. Based on the GPS data recorded during sample collection,
the sample collection sites are located between 12O28.0988’-
13O19.9522’N and 38O53.1815’- 39O40.9870’E with an altitude
range of 1342-1822m a.s.l.
Sample collection
A total of 93 sorghum roots with adhering soil samples were
collected in sterile plastic bags. Samples were collected based
on altitude differences of sorghum plant growing areas, cultivar
types and plant age group. At each sampling site, plant roots with
adhering soil (approximately 50g) were uprooted and placed
into a sterile plastic bag. Care was taken to keep rhizosphere soil
intact around the root. The collected samples were kept in ice-box
and transported to Ethiopian Biodiversity Institute Microbiology
Laboratory. All samples were kept at 4 ˚C until use [22-24].
Isolation of Rhizobacteria
Sorghum roots with adhering soils were merged into 17
composite samples separately based on similarity of cultivar
type, plant and age group. The root adhering soils were dislodged
from the roots using sterile distilled water by shaking at 250rpm
for 20 minute and the root washing solutions were used for the
isolation of rhizoplane bacteria [25]. For the isolation of bacterial
endophytes, merged and washed roots were surface sterilized in
99% ethanol for 1min, 3% NaOCl for 6 minutes, and 99% ethanol
for 30 seconds and followed by rinsing with sterile distilled water
for 6 times [23]. Before homogenization, a root fragment was
imprinted on nutrient agar to serve as a sterility check. Roots were
homogenized and macerated with a sterile mortar and pestle [26].
The root washing solutions and homogenized roots were serially
diluted (10-2 to 10-4) aseptically for inoculation. 0.1ml inoculums
of the prepared samples were spread onto Nutrient agar plates
and incubated at 30+2 ˚C for 48h [27,28]. Bacterial colonies with
distinct and peculiar morphologies were selected and re-streaked
to obtain pure colonies [24].
In vitro screening of bacteria for P-solubilization potential
Phosphate solubilization ability of the isolated bacteria was
determined on Pikovskaya’s agar. The isolates were spotted
onto Pikovskaya’s agar and incubated for 7 days at 30 ± 2 ˚C. The
presence of halo zone around the bacterial colony was considered
as indicator for positive phosphate solubilization. Further, the
solubilization index (SI) of the isolates was determined by
measuring the halo zone of clearance (HD) in the Pikovskaya’s
agar plates and the colony diameter (CD) [29]. SI was calculated
with the formula: SI = (CD+HD)/CD. Three replicate plates were
used for each isolate [30].
Identification of P-solubilizing rhizobacteria
Preliminary identification of P-solubilizing Rhizobacteria
isolates were performed by examination for cell morphology
using optical microscopy, Gram staining, and colony morphology
[27,24]. Biochemical identification including the carbohydrate
fermentation patterns and chemical sensitivity tests were
determined using GEN III Biolog bacterial identification system
kit. The Biolog GEN III Micro Plate analyzes a microorganism
in 94 phenotypic tests: 71 carbon source utilization assays
and 23 chemical sensitivity assays. The test panel provides a
“Phenotypic Finger print” of the microorganism that can be used
to identify it at the species level. The plates contained 96 wells,
with a dehydrated panel of necessary nutrient medium (a carbon
source), biochemical and tetrazolium violet. Tetrazolium violet is
a purple formazan, a redox dye that turns purple when reduced,
indicating use of the carbon source provided or resistance to
inhibitory chemicals. Each plate contained a positive and negative
control well. Pure culture of bacteria isolates was grown on Biolog
BUG agar plates at 30 ± 2 ˚C for 20-24 hours. Single colonies were
swabbed and suspended in inoculating fluid A. Cell suspensions
(100μl) adjusted at 90-98% transmittance was pipetted into 96
well Biolog Micro-plates for carbon utilization and chemical test.
Panels were incubated at 30 ± 2 ˚C for 20-24 hours. The microplates
were inserted into the Omnilog automatic system and
the identification process was carried out using GEN III Biolog-
Omnilog identification system software [31].
Data analysis
Data were analyzed using SPSS software version 20 (SPSS
Inc., Chicago, IL, USA). Coefficient of variation was calculated for
the significances of differences within samples and ANOVA was
employed for significances of differences between mean counts
of microbial groups. DIVA_GIS 7.5.0 was used for mapping study
areas.
Results and Discussion
In vitro screening of P-solubilizing rhizobacteria
Ninety-four bacteria were isolated from root washing
solutions and surface sterilized roots on nutrient agar. Fifty-one
bacteria were isolated from sorghum root washing solutions
which were prepared from the root adhering soils and the rest
43 were endophyte bacteria isolated from sorghum roots. These
94 bacterial isolates were evaluated for their ability to solubilize
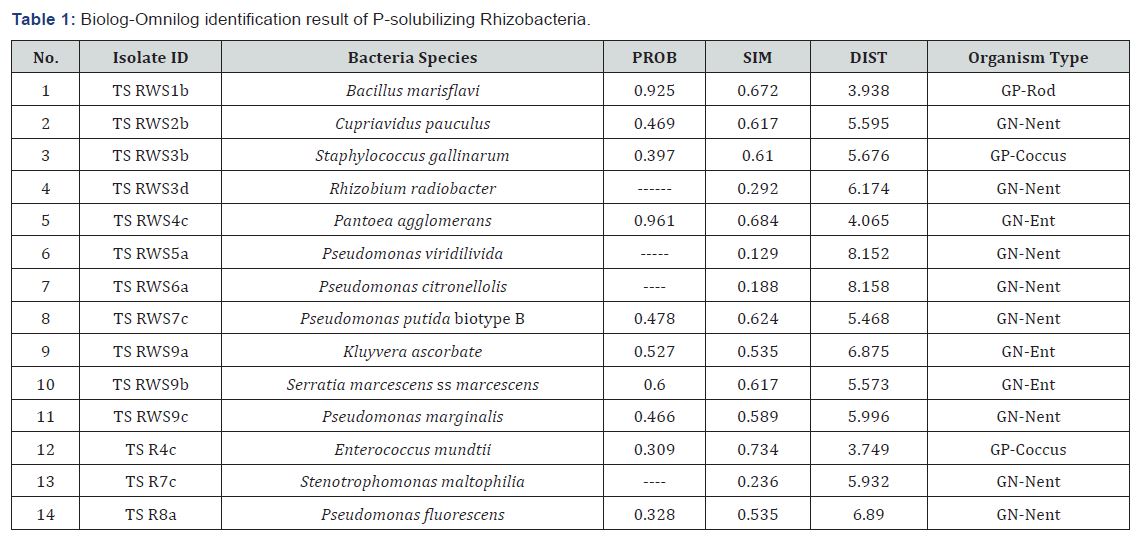
phosphates on Pikovskaya’s agar plates (Table 1). Fifty four of the
94 (57.5%) rhizobacterial isolates showed clearly visible haloes
(>0.50cm) around their colonies on Pikovskaya’s agar after seven
days of incubation. The solubilization index (SI) of the potential
P-solubilising rhizobacterial isolates differed significantly
(p<0.05) and ranged from 0.5 to 4.83. Bacterial strain TS RWS7b
produced the largest zone of solubilisation, followed by TS RWS
1b.
Identification of P-solubilizing rhizobacteria
Based on colony morphology shown on nutrient agar and
Biolog Universal Growth (BUG) agar, and Gram staining similarity,
the 54 P-solubilizing Rhizobacteria screened from root washing
solutions and sorghum roots were clustered into 17 representative
isolate morphological groups. Inoculums of the 17 clustered
representative isolates were prepared and transferred into GEN
III Micro-plates. After 24 hours of incubation at 30+2 ˚C, the microplates
were subjected to Biolog-Omnilog bacterial identification
system test. Fourteen of the 17 clustered representative
P-solubilizing Rhizobacteria isolates were identified (Table 1).
Eleven of the 14 identified P-solubilizing Rhizobacteria were
isolated from root washing solution and the rest 3 were isolated
from sorghum root. Gram negative rhizobacteria dominated
the system accounting for 78.57% (11/14) of the identified
P-solubilizing Rhizobacteria isolates (Table 1,2). Previous
observation showed that the rhizosphere of many agriculturally
important plants favors more Gram negative rhizobacteria than
the Gram positives [4,32]. The largest solubilization index was
also produced by Gram negative isolate when compared with
Gram-positive isolate. Some of the isolates lost their capacity for
phosphate solubilization on repeated sub-culturing as previously
reported in many other studies [33,34].
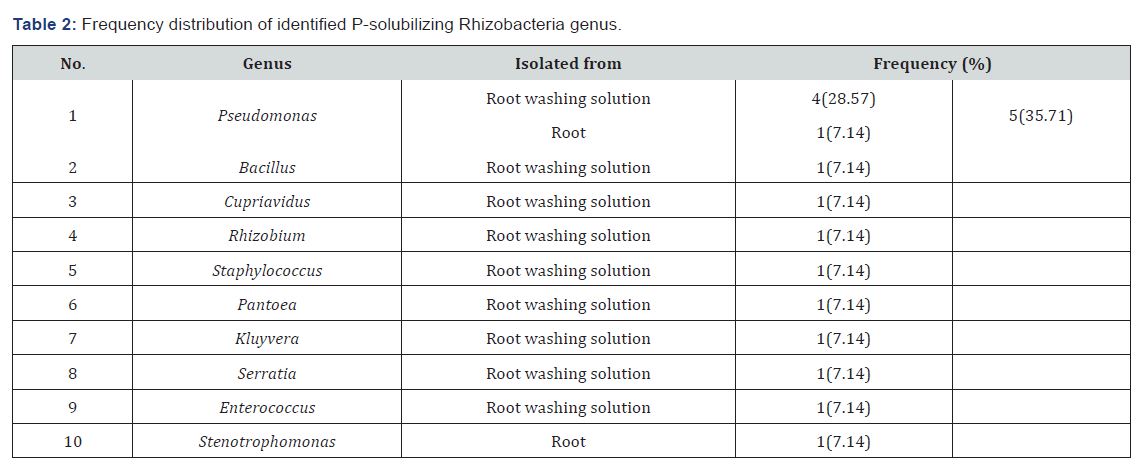
Ten different genera of Rhizobacteria were identified. Most of
them were isolated from root washing solutions. Eight of the 10
identified Rhizobacteria genera were isolated only from sorghum
root washing solutions. But, only Stenotrophomonas species
was isolated from root. Meanwhile, Pseudomonas species was
isolated from both root washing solutions and root. Members of
the phosphobacteria were dominated by the genus Pseudomonas
(35.71%) (Table 2). Pseudomonas are the most dominant genera
commonly reported in many plant studies [35].
Conclusion
This study showed that there are a large proportion of P-solubilizing
rhizoplane and endophytes rhizobacteria associated with
Sorghum bicolor L. Pseudomonas is the most dominant rhizobacteria
both in the root adhering soil and roots of sorghum. In general,
Gram negative bacteria were not only more predominant than
Gram positive bacteria but also, they produced the largest solubilization
index. This finding indicated that there is a great number of rhizobacterial potential associated with Sorghum bicolor L. which
can be utilized for development of P-solubilizing bio-fertilizers.
To know more about Journal of Agriculture Research- https://juniperpublishers.com/artoaj/index.php
To know more about open access journal publishers click on Juniper publishers



Comments
Post a Comment